Unpublished Work 1: Galactose Memory and Yeast Surface Display
In an effort to not waste the taxpayer dollars which fed this work, and to not let good knowledge go to waste, I want to put up some little research stories which produced true information but didn’t clear the threshold for a paper. This will be the first of 3 or 4, written informally and without perfect completeness in mind. If anyone is interested in more details, feel free to reach out.
This side quest stemmed from an observation I made about one of my colleagues work. His system to evolve nanobodies, which relies on OrthoRep and yeast surface display, had a low display rate. For a little context; yeast surface display is a method of attaching antibodies, nanobodies, enzymes, etc to the yeast cell surface, and is commonly used for screening of binding proteins. Often, it uses a cell wall bound protein (GPI) or the yeast mating architecture (Aga1/2).
The system was built to turn on display when the sugar in the yeast growth media is swapped from glucose to galactose but only about 10-20% of cells containing the galactose inducible nanobody would induce, and it would be a bimodal distribution of induced / noninduced.
His system relies on rounds of in-vivo mutation by OrthoRep, and FACS based sorting of improved binders. I noticed that after a couple of rounds of induction and sorting that the display level would increase substantially, from ~15% to 60%. The postdoc who built this system reasoned that it was because the in-vivo mutation system / sorting was enriching better displaying nanobodies.
I disagreed and went to test the effects of repeated rounds of galactose induction without any sorting or hypermutation, just a CEN/ARS plasmid getting induced multiple times. I found that multiple consecutive rounds of galactose induction, so gal, gluc, gal, gluc, gal, etc, would increase the fraction of cells which are displaying the nanobody each round, and the effect was not related to properties of the nanobody itself.
This was a bit of coup for me as it was within the first few months of my PhD. When I presented it to a multi university research consortium doing similar work Prof Aaron Ring from Yale stopped the meeting to give me a round of applause and said “I’ve been working with yeast for 20 years and never realized this.” That was nice.
What I had accidentally rediscovered was a phenomenon called “galactose memory.” The circuit that allows yeast to kick on the galactose sugar response, and thus drive the surface display architecture, has an activation cascade which becomes easier to trigger as some components aren’t fully reset between activation rounds. The precise mechanism is pretty well studied, though there’s a bit of a debate about whether it’s fully protein based, or if there’s an DNA based component.
This info is useful for people doing these sorts of antibody screens, as the initial sorting for binders can be demanding for a sorter. Say you have a library of 10 million sequences, and can sort 2,000 cells per second, covering the library once will take 13.8 hours at 10% display, or 2.2 hours at 60% display.
To attempt to make it more useful I aimed to exploit the known mechanism of memory and make a cell line with a slightly modified circuit for gal induction, which would mimic gal memory at the first induction. The aim was to discover a way to simply modify library destination strains to have the improved display property without having to do multiple rounds of gal induction.
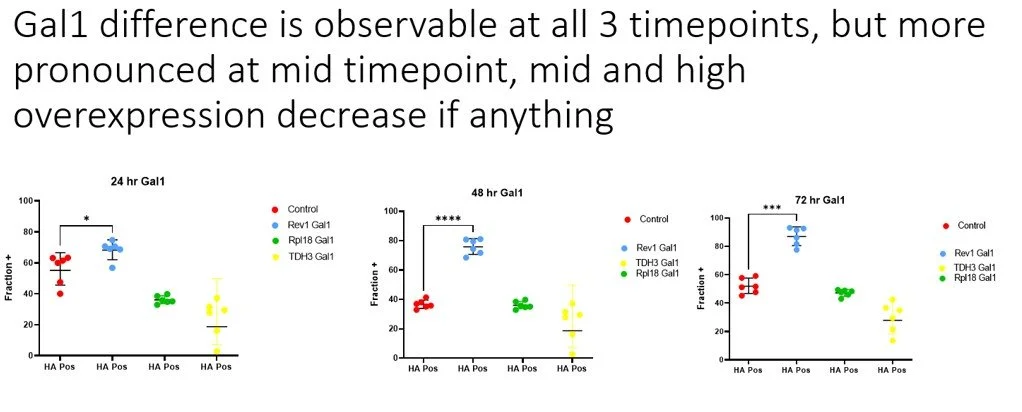
This is where things got weird. I found that overexpressing one element (gal1) of the galactose memory pathway would increase the induction dramatically compared to no overexpression, but only when the genomic integration Gal1 and nanobody plasmid were co-transformed. If the integration cassette was introduced prior to the transformation of the nanobody construct, then it would not increase display. I also tested Gal 2, 3, 4, 10, with the same set of 3 promoters, and those did not increase display.
The nuances of why, I don’t know. This was about the point where I moved on, working out the precise dynamics of gene expression that would explain this behavior would probably not be worth it. It could be a very deep rabbit hole, and unless it results in a cell line with improved display properties without co-transformation it may not be terribly useful, as co-transformation of a second plasmid along with the nanobody library could limit the transformation efficiency (though in hindsight I think this was a bad assumption because I later found competent yeast can easily take up multiple plasmids). It was a shame to not get to publish this finding in a journal, but sometimes the juice isn’t worth the squeeze.